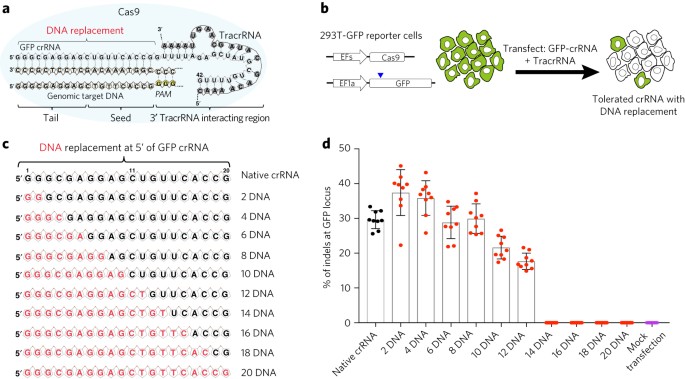
(A) Binding sites of amiRNAs in different regions of sgRNA Region 1 represents the spacer sequence, and regions 2 and 3 are located in the sgRNA backbone (B) Effects of amiRNAs on the expression of naked sgRNA by qRT PCR GAPDH was used as a control Data are the mean ± SD from five experimentsEncodes sgRNA targeting position 6 in pColE1_70a_deGFP_KanR with one mismatch in the nonseed region Depositor Vincent Noireaux Insert sgRNA targeting position 6 in pColE1_70a_deGFP_KanR, with one mismatch in the nonseed region Use Crispr s Expression Mutation Promoter J Availability Unlike the proximal region, the PAMdistal region has been considered less important in determining the sgRNA specificity However, in our study, we show that this region may actually contribute

Sequence Recognition And Structure Of Synthetic Crispr Rna A Download Scientific Diagram
Seed region sgrna
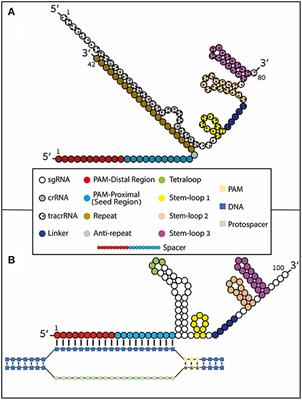
Seed region sgrna-The use of CRISPR–Cas9 as an RNAprogrammable DNA targeting and editing platform is simplified by a synthetic singleguide RNA (sgRNA) mimicking the natural dual trans activating CRISPR RNA (tracrRNA)–CRISPR RNA (crRNA) structure This review aims to provide an indepth mechanistic and structural understanding of Cas9mediated RNAguided Small interfering RNA (siRNA) is also known as short interfering RNA or silencing RNA In the literature, synthetic siRNA constructs are generally denoted by "gene name" siRNA (eg p53siRNA), with constructs targeting the same gene distinguished by the addition of numbers after the construct name (eg p53siRNA1 and p53siRN)



2
Of note, both motifs are located in the sgRNA "seed" region (the PAMproximal 10–12 bases of the targeting sequence) that is important for pairing with the target DNA (Jinek et The argininerich bridge helix and the immobilized seed region of sgRNA plays key role in the initiation of Rloop To examine the detailed requirement of the seed region in protospacers, consecutive dinucleotides transversion mismatch sequences with 1 nt step size of target DNA were designed to study the tolerance of SpyCas9 for mismatched Several previous studies show that mismatches in the seed region (the 10–12 base pairs adjacent to the PAM) are critical and determine the specificity of Cas9 compared to the distal part of the sgRNA sequence (nonseed sequence) (Hsu et al, 13;
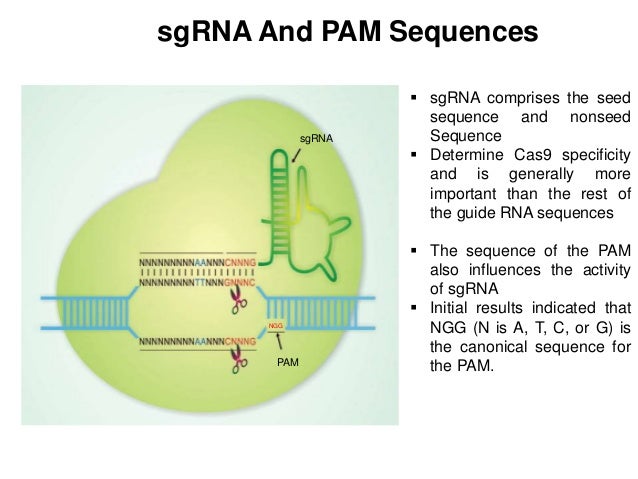
(only NGG PAM, only G as 5' base, offtarget tolerates many mismatches and ignores nonseed region, introns, purpose is knockout (only first 3 coding exons are allowed) and UTRs are excluded) Single design Paired designs Start sgRNA search Reset formThe seed region, the Cas9RNP can still bind sites with some seed region mismatches More recently, Refolded sgRNA was aliquoted and stored at 80⁰C To assemble the Cas9RNP, 3 µM of purified Cas9 was incubated with 36 µM refolded sgRNA at 37⁰C for 10 minutes ( mM HEPESKOH, pH 75, 150 mM KCl, 10% Glycerol, 1 mM MgClDefining the seed sequence of the Cas12b CRISPRCas effector complex tospacer region was performed with Bth Lib F (5 15 nM sgRNA, 1 nM beacon and 50 nM target DNA competitor
Upon being complexed with sgRNA, SaCas9 confers stable binding to the DNA target when 6 PAMproximal matches exist It then triggers sequential DNA unwinding from the PAMproximal region to the PAMdistal end and samples adjacent to the DNA for guide RNA complementarity The seed sequence influences the specificity of Cas9sgRNA binding through multiple potential mechanisms The sequence of the seed region determines the frequency of a "seed NGG" in the genome, and controls the effective concentration of the Cas9sgRNA complex (Cas9 binding or sgRNA abundance and specificity) 21, 25 Meanwhile, Urich seeds are likely toThe seed sequence influences the specificity of Cas9sgRNA binding through multiple potential mechanisms The sequence of the seed region determines the frequency of a "seed NGG" in the genome, and controls the effective concentration of the Cas9sgRNA complex (Cas9 binding or sgRNA abundance and specificity)21,25 Meanwhile,
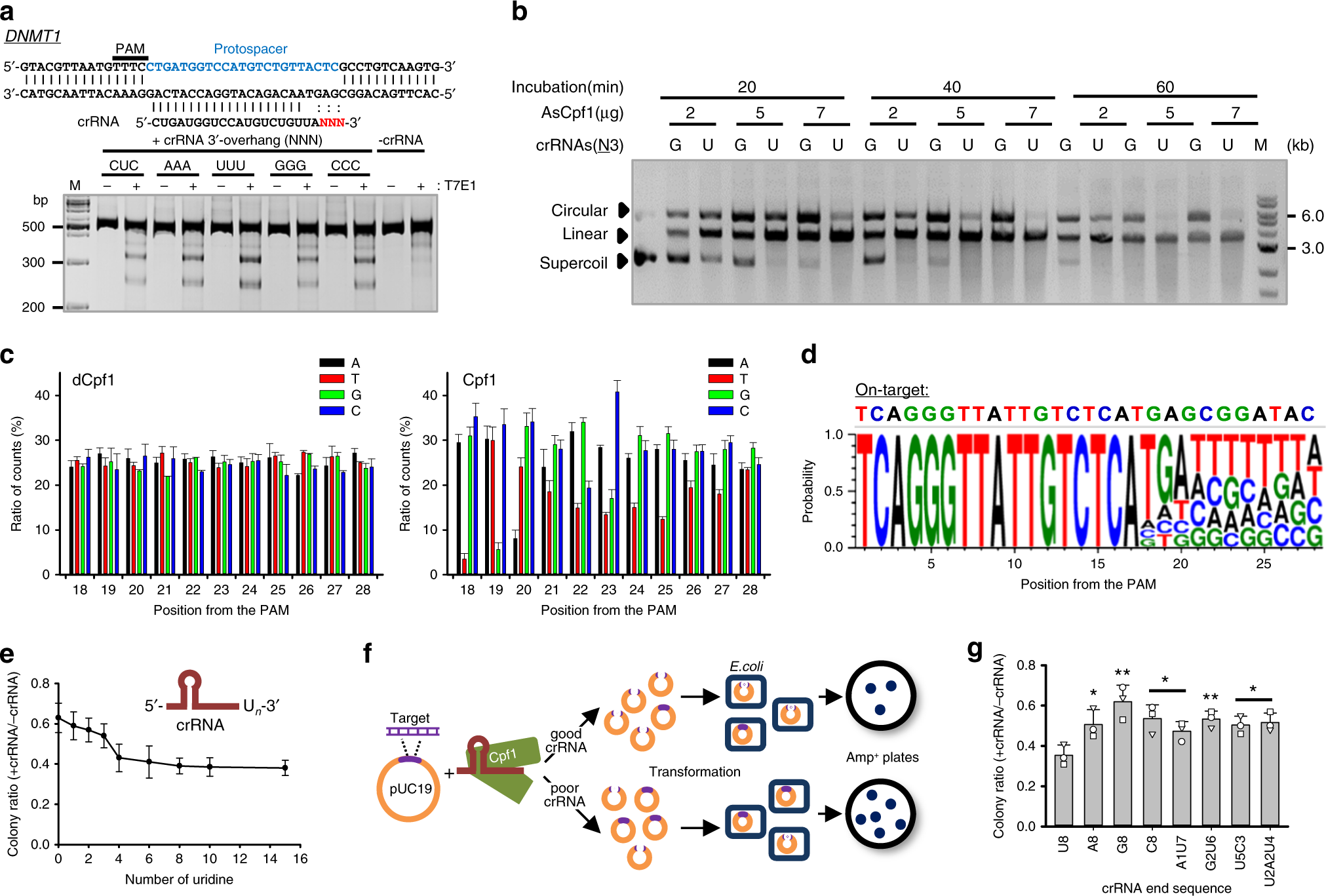



Highly Efficient Genome Editing By Crispr Cpf1 Using Crispr Rna With A Uridinylate Rich 3 Overhang Nature Communications




Jgqkaqhmf1 92m
SgRNA Complementarity in the Seed Region Is Critical for Cas9 Cleavage of Nucleosomes—To systematically screen how sgRNA mismatches impact Cas9 activity on nucleosomes, we generated a panel of sgRNA mutants containing single mismatches at locations throughout the sgRNA guide segment (Fig 2A) Each mutant sgRNA was tested for its activity inWhile these effects can be deduced to the sgRNA expression level or stability, the general pattern of less important seed region in eSpCas9 may suggest that the mutations partially reprogram the recognition or subsequent interaction and cleavage function of the eSpCas9–sgRNA complex with respect to its DNA substrate, possibly because of the Every backbone phosphate of the seed nucleotides at positions 2–8 from the anchored 5′terminal nucleotide preordered on the AGO protein to make stable basepairing between the siRNA seed region and target mRNA in an Aform helix 11,12 The efficiency of the offtarget effect is positively correlated with the thermodynamic stability of the




Single Base Resolution Increasing The Specificity Of The Crispr Cas System In Gene Editing Molecular Therapy
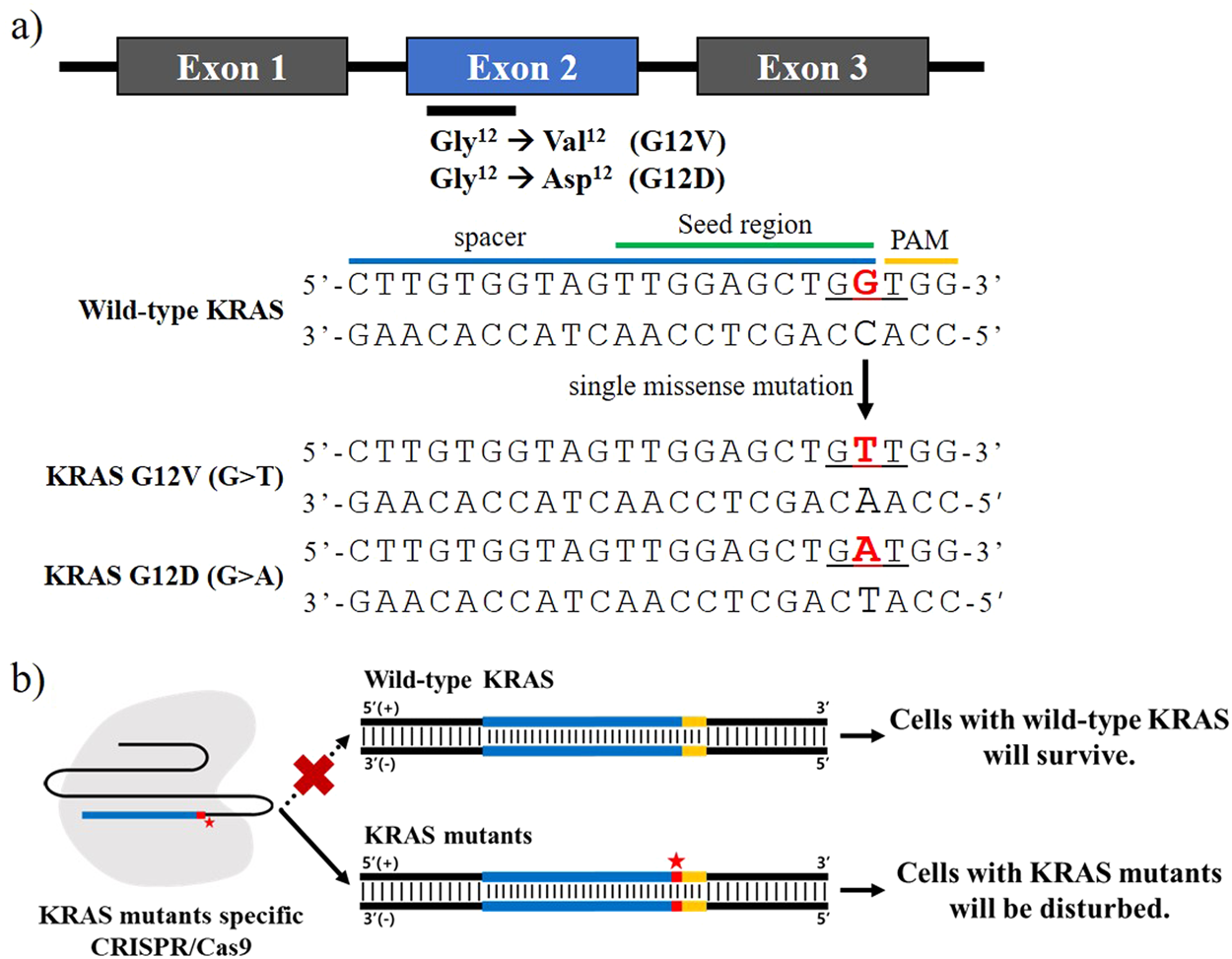



Selective Targeting Of Kras Oncogenic Alleles By Crispr Cas9 Inhibits Proliferation Of Cancer Cells Scientific Reports
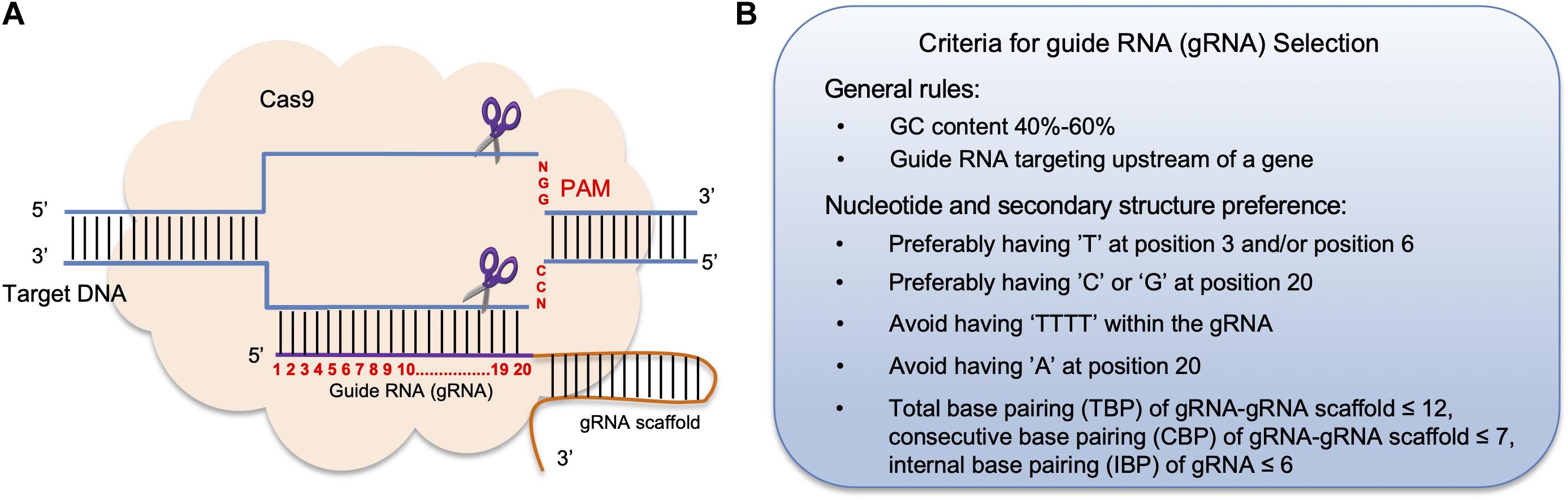
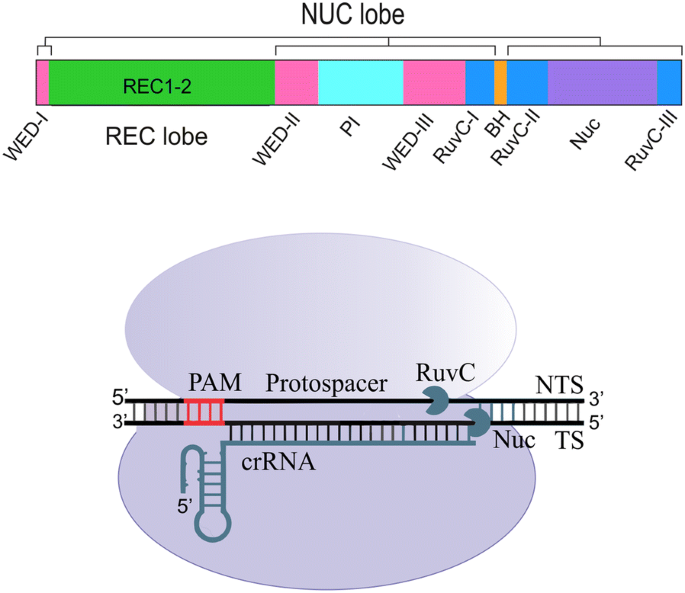
Target DNA, the Nme1Cas9 sgRNA seed is preordered in a nearly Aform conformation, similar to that of the sgRNA of SpyCas9 (Jiang et al, 15) The sugarphosphate backbone of the ordered seed region forms extensive interactions with the argininerich BH Only 13 bp of the repeatantirepeat duplex contact the REC1 However, when designing the proximal sgRNA sequence of PAM, guanine is preferred in the first base of the seed region, cytosine is avoided, because guanineenriched sgRNA is easier to fold, forming a more stable structure and reducing offtarget effect The length of sgRNA is also closely related to its specificity In recent years, the type II eukaryotic cluster regularly spaced palindromic repeat sequence (CRISPR) system has been applied to rapidly expanding eukaryotic hosts as an efficient tool for targeted genome editing, regulation and labelingIn these systems, a complex of Cas9 proteins and a pair of RNA recognizes a DNA target consisting of a short piece of complementary DNA at the




Modulation Of Binding Affinity And Specificity By Guide Rna Variants Download Scientific Diagram
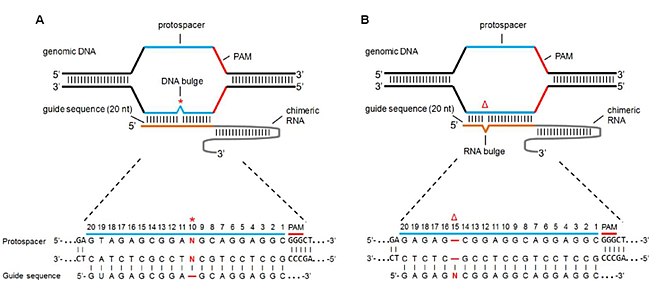



Off Target Genome Editing Wikipedia
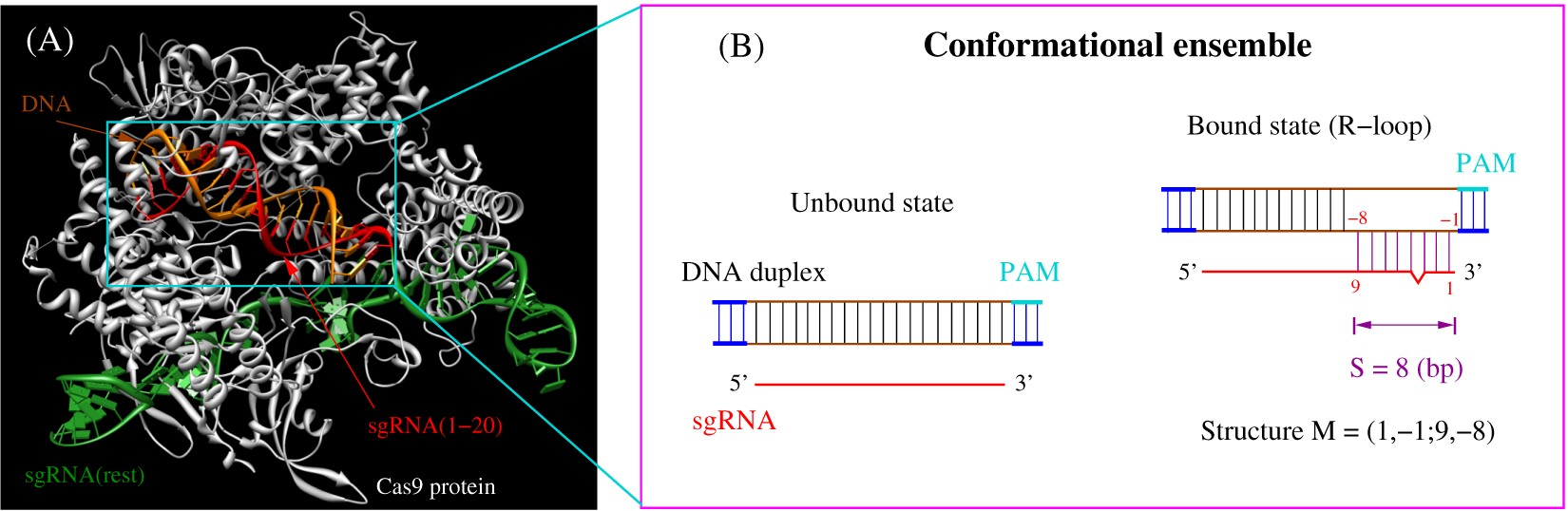
RNA (sgRNA) to cleave specific DNA sequences within eukaryotic cells, which facilitates its use asatoolforgenomeengineering(3–5)Biochemical and genome occupancy studies have established the PAM and adjacent ~5 to 8 base pairs (the "seed" region) of the DNA target site as the basis for Cas9 DNA interrogation and offtarget Each mutant sgRNA was tested for its activity in directing Cas9 cleavage of naked DNA or nucleosome substrates, following a 30min incubation A number of single sgRNA mismatches had significant effects on cleavage of the naked DNA substrate, particularly mismatches in the sgRNA seed region (Fig 2, B and C), consistent with previous studies (3 Crystal structures of Cas9 bound to sgRNA and a target DNA strand, with or without a partial PAMcontaining nontarget strand, show the entire nt guide RNA segment engaged in an Aform helical interaction with the target DNA strand (12, 13) How the "seed" region within the guide RNA specifies DNA binding has remained unknown



2




Addgene Crispr Guide
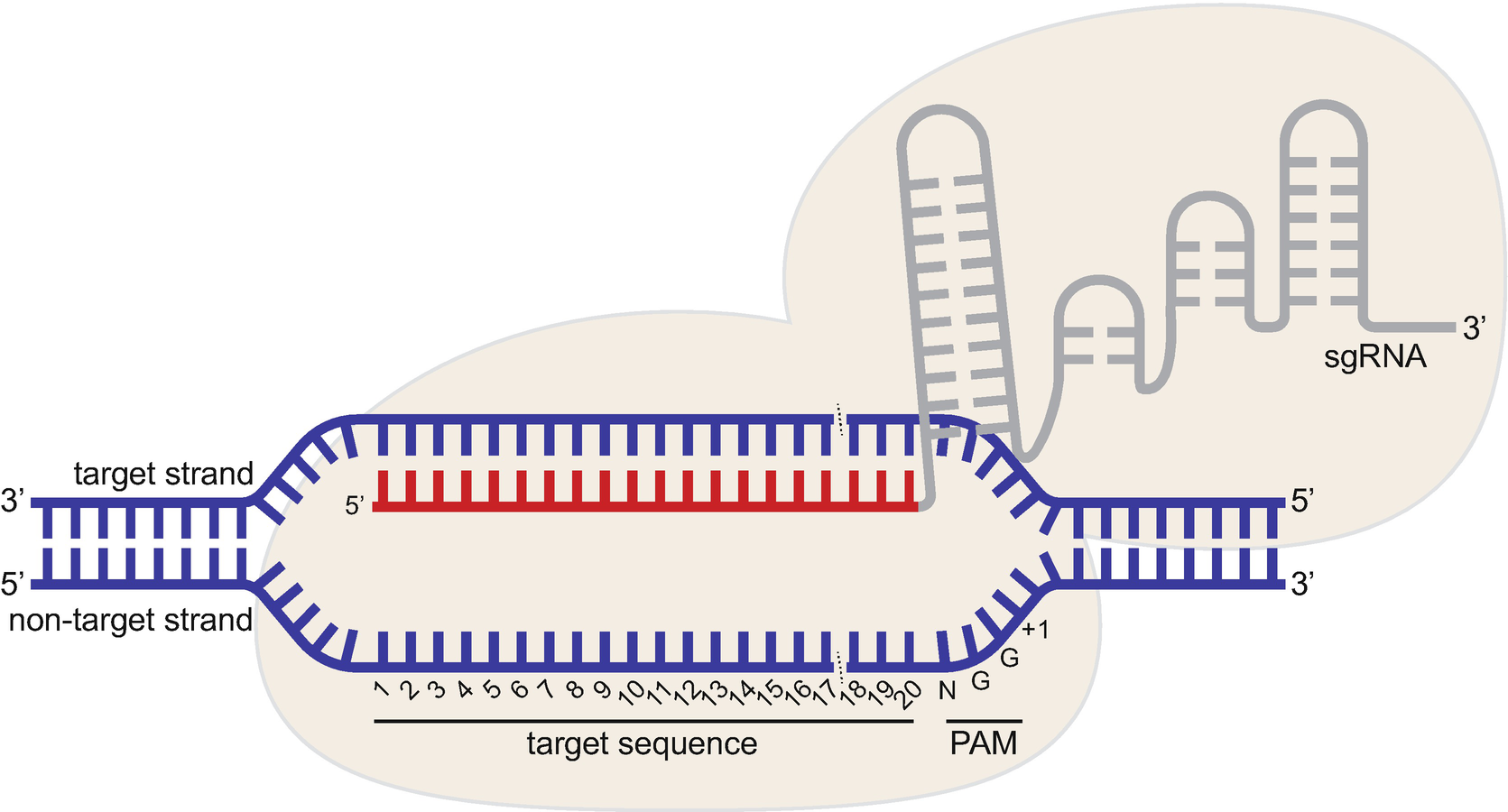
Importantly, the spacer region of the gRNA remains free to interact with target DNA Cas9 will only cleave a given locus if the gRNA spacer sequence shares sufficient homology with the target DNA Once the Cas9gRNA complex binds a putative DNA target, the seed sequence (810 bases at the 3′ end of the gRNA targeting sequence) will begin toNNGRRT (R = A or G) Custom PAM Method for determining offtargets in the genome Offtargets with up to mismatches in protospacer (Hsu et al, 13) Offtargets may have no more than mismatches in the protospacer seed region (Cong et al, 13) Efficiency score Doench et al 14 only for NGG PAMWe therefore define position 15 as the 'seed' region of the sgRNA For Nanogsg2, the guide match extends to about 1012 bases to the 5′ end, possibly
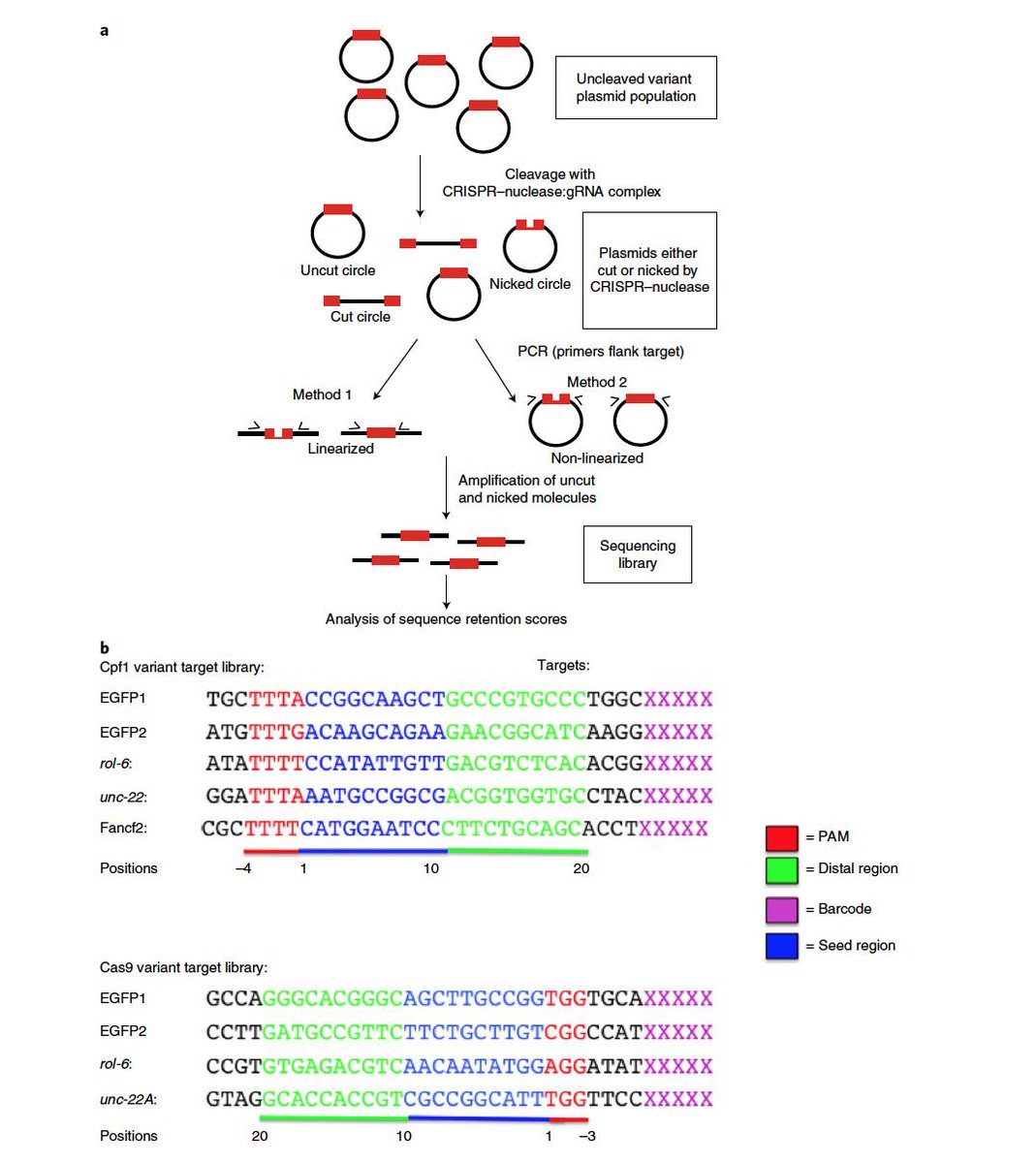



Dr Gaetan Burgio Md Phd Interesting This Paper Published Today In Naturemicrobiol Shows That Crispr Cas9 Or Cas12a Cpf1 Can Display An Inherent Potent Nicking Activity In Yeast This Nicking




Sgrna Multiplexing Strategies A Rna Endonuclease Csy4 Recognizes A Download Scientific Diagram
Since the seed region of gRNA is imperative for the activity, therefore, we calculated the GC content percentage of the PAM proximal seed region (1–12 nt) and the PAM distal region (13– nt) In the case of the PAM proximal seed region (1–12 nt), the GC content positively and significantly impacted the cleavage efficacy (pvalue = 32EBacterial type II CRISPRCas9 systems have been widely adapted for RNAguided genome editing and transcription regulation in eukaryotic cells, yet their in vivo target specificity is poorly understood Here we mapped genomewide binding sites of a catalytically inactive Cas9 (dCas9) from StreptococcDefine positions 1–5 as the 'seed' region of the sgRNA For Nanogsg2, the guide match extends to about 10–12 bases to the 5′ end, possibly due to the presence of multiple U's in the seed that lowers the thermodynamic stability of the sgRNADNA interaction For Nanogsg3 and Phc1sg2, an exact match to the 5nucleotide seed followed by




Computational Approaches For Effective Crispr Guide Rna Design And Evaluation Sciencedirect



2
Within the seed region of sgRNA5, position 5, 6 (core sequence) and 3, 9 (seed region but not core sequence) were randomly chosen to generate all possible single nucleotide mismatches, whileWe therefore define position 15 as the 'seed' region of the sgRNA For Nanogsg2, the guide match extends to about 1012 bases to the 5′ end, possibly due to the presence of multiple Us in the seed that lowers the thermodynamic stability of the sgRNADNA interaction For Nanogsg3 and Phc1sg2, exact match to the 5 The effect of a single mismatch is more pronounced for mismatches far away from the PAM site (PAMdistal region) as opposed to PAMproximal nucleotides, sometimes referred to as the seed region Mismatch tolerance appears to be maximal at the th position This suggests that guides should be carefully designed to avoid mismatch at that position
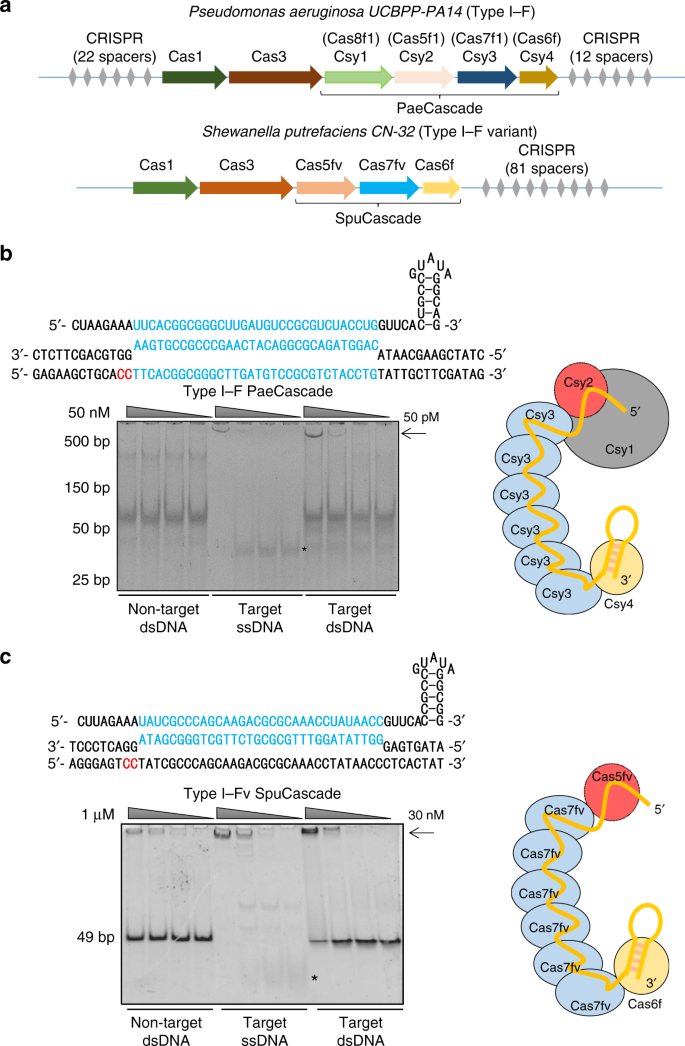



Repurposing Type I F Crispr Cas System As A Transcriptional Activation Tool In Human Cells Nature Communications




A Model For Cas9 Target Binding And Cleavage A In The Unbound State Download Scientific Diagram
Mismatches between the target and CRISPR RNA guide outside the seed have minor effects on target binding, thus contributing to offtarget activity of CRISPRCas effectors Here, we define the seed sequence of the Type V Cas12b effector from Bacillus thermoamylovorans While the Cas12b seed is just five bases long, in contrast to all otherThe CRISPRassociated protein is a nonspecific endonuclease It is directed to the specific DNA locus by a gRNA, where it makes a doublestrand break There are several versions of Cas nucleases isolated from different bacteria The most commonly used one is the Cas9 nuclease from Streptococcus pyogenes Figure 1 For example, a mismatch in the seed region may cause a notable reduction of the cleavage activity of Cas9/sgRNA, while mismatches in the other regions of the protospacer (the nonseed region) have a much weaker effect
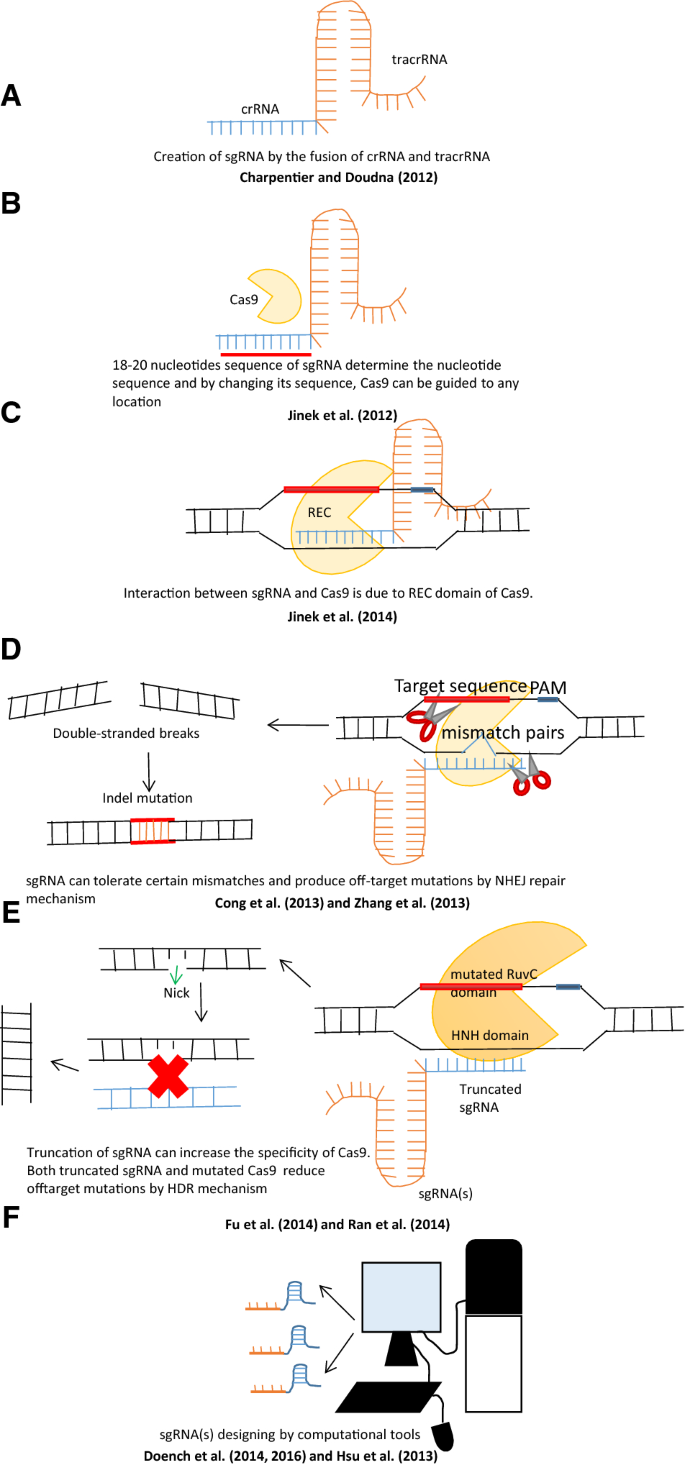



An Overview Of Designing And Selection Of Sgrnas For Precise Genome Editing By The Crispr Cas9 System In Plants Springerlink




Target Dna Cleavage By Crispr Cas12a Cpf1 Using Chimeric Dna Rna Download Scientific Diagram
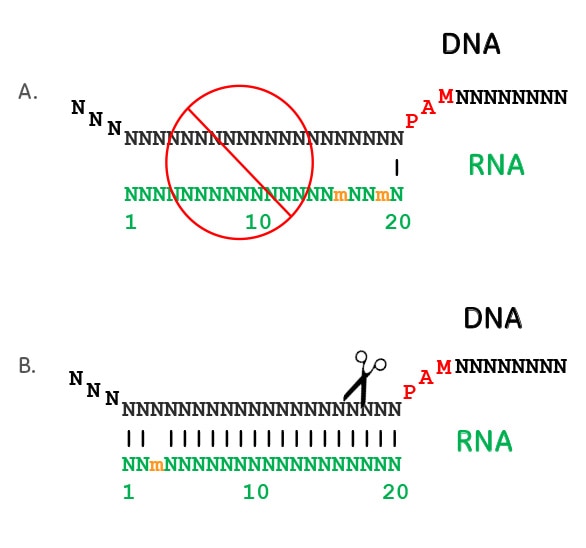
Seed region The chemical modifications of deoxyribonucleic acid (DNA), PS, and DNA−PS were introduced into all of the seven nucleotides of the seed region of the siRNA guide strand (Figure 2b) In addition, 3 (positions 4−6), 5 (positions 3−7), Use the Basic Local Alignment Search Tool (BLAST) to check the specificity of sgRNA binding in the genome To ruleout additional binding sites and offtarget effects, BLAST search the 14nt specificity region, which includes the 12nt 'seed region' of the sgRNA and 2nt of the 3nt PAM region, in the genome of interest Genomic regions that have at least 11nt homology to the sgRNA seed region and a neighboring PAM sequence were sequenced from F1 whiteeyed flies generated by the optimized system Mismatches between the potential offtargets and the targeted region are underscored The PAM sequences are in bold type
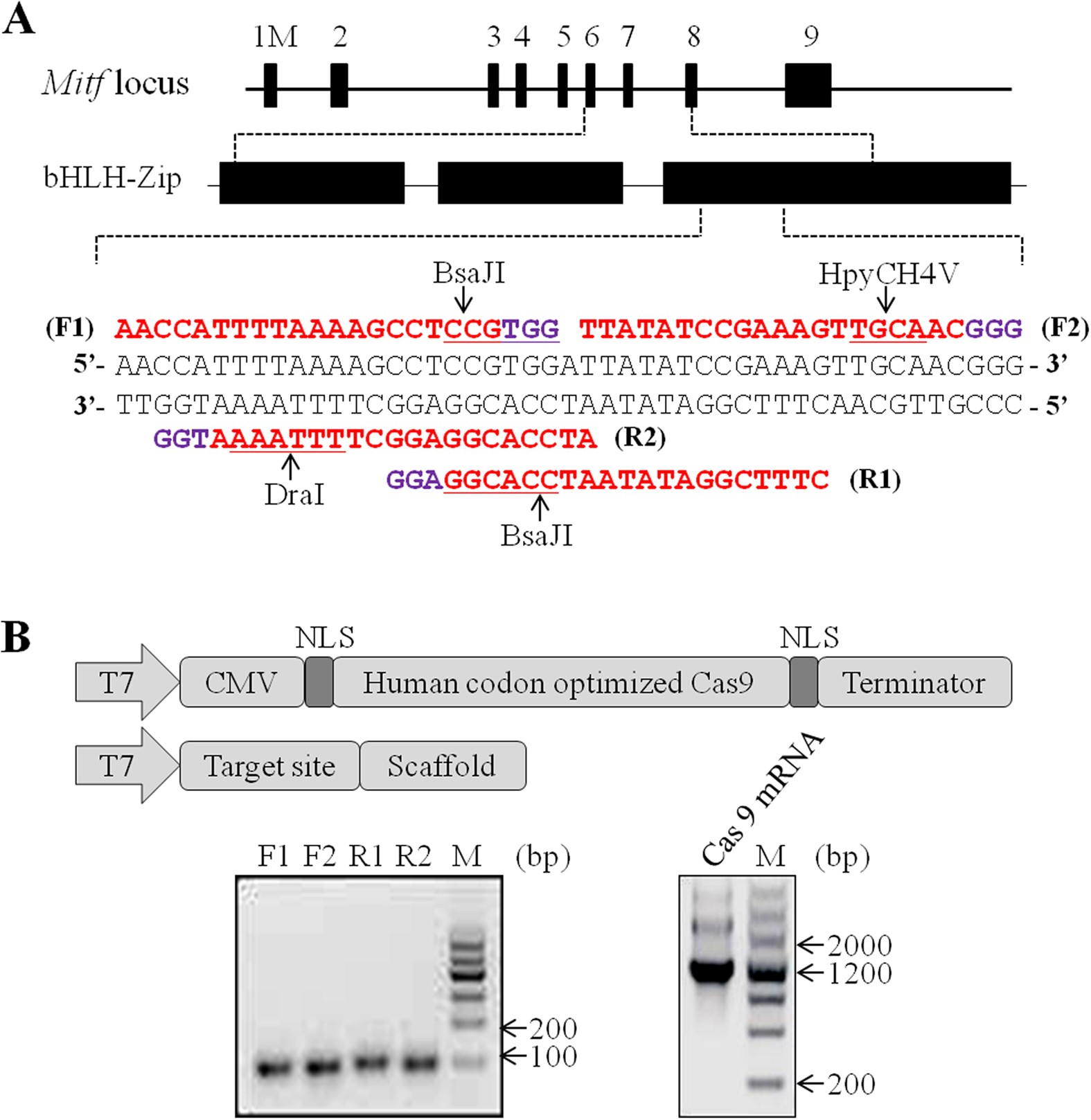



Efficient Crispr Cas9 Mediated Biallelic Gene Disruption And Site Specific Knockin After Rapid Selection Of Highly Active Sgrnas In Pigs Scientific Reports



Full Article Defining The Seed Sequence Of The Cas12b Crispr Cas Effector Complex
UUU was significantly absent in the seed region of functional gRNAs compared with that in nonfunctional gRNAs (08 % versus 84 %, P = E–7) Overall nucleotide usage Within the nucleotide gRNA sequence, the average counts for adenine were 46 and 33 for functional and nonfunctional gRNAs, respectively ( P = 93E–18)An exact match to 3′ half of the sgRNA (known as the seed region) followed by a PAM motif, may be a target for offtarget activity If there are mismatches in the 3′ half of the guide RNA, or if the PAM is absent, then the sequence is unlikely to be cleaved Do note, however, that this is extrapolation from research in mammalian systemsFor example, we found that two mutations in the seed region can increase U6 promoter transcribed sgRNA's abundance by at least 7 fold 2) Changes that alter the number of seedmatching sites in the genome, which can vary by 100fold
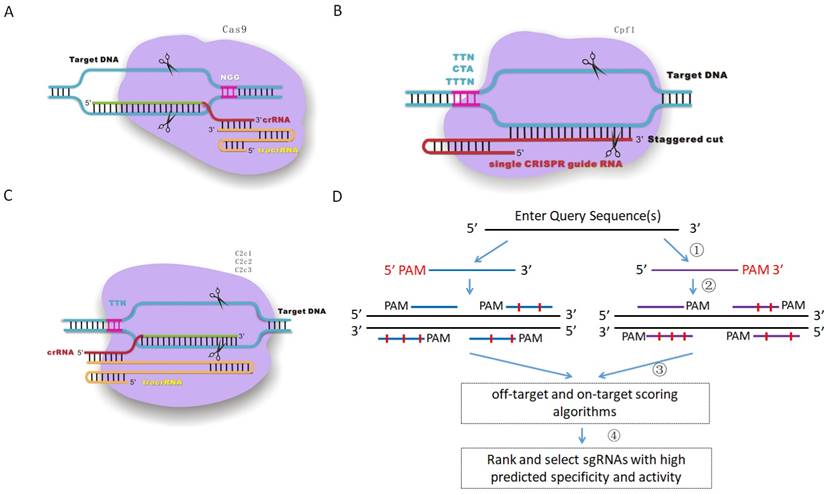



Crispr Offinder A Crispr Guide Rna Design And Off Target Searching Tool For User Defined Protospacer Adjacent Motif




Genes Free Full Text Spcas9 And Lbcas12a Mediated Dna Editing Produce Different Gene Knockout Outcomes In Zebrafish Embryos Html
Perfect match to the 12nt seed region of the sgRNA and 3 mismatches in the outer segments M4_POT Perfect match to the 12nt seed region of the sgRNA and 4 mismatches in the outer segments M5_POT Perfect match to the 12nt seed region of the sgRNA and 5 mismatches in the outer segments Total_Noof_POTCRISPRCas9 is quickly revolutionizing the way we approach gene therapy CRISPRCas9 is a complexed, twocomponent system using a short guide RNA (gRNA) sequence to direct the Cas9 endonuclease to the target site Modifying the gRNA independent of the Cas9 protein confers ease and flexibility to improve the CRISPRCas9 system as a genomeediting tool gRNAs have been This is a major improvement over the proteinbased gates, which have problems in all of these areas The "operator" that is bound by the sgRNA seed region is also relatively small (13 bp) and can be easily inserted between the 10 and 35 region of a promoter (TetR homologue operators range from to 50 bp)




Improving Crispr Genome Editing By Engineering Guide Rnas Trends In Biotechnology




The Crispr Cas9 System For Plant Genome Editing And Beyond Sciencedirect
Cas9sgRNA complexes are able to load onto multiple sites with short seed regions adjacent to 5′ NGG 3′ protospacer adjacent motifs (PAM) Yet among 43 ChIPseq sites harboring seed regions analyzed for mutational status, we find editing only at the intended ontarget locus and one offtarget siteTions of a ''seed'' region Our results provide a quantitative basis for the DNA cleavage patterns measured in vivo and observations of greater reported target specificity for Cas12a than for the Cas9 nuclease INTRODUCTION CRISPRCas systems are revolutionary new tools for gene editing applications (Hsu et al, 14) The nucleaseCRISPRCas9 is a simple twocomponent system thatThe seed sequence is essential for the binding of the miRNA to the mRNA The seed sequence or seed region is a conserved heptametrical sequence which is mostly situated at positions 27 from the miRNA 5´end Even though base pairing of miRNA and its target mRNA does not match perfect, the "seed sequence" has to be perfectly complementary




1 Primary Structure Of Sgrna The Sgrna Contains Three Regions Variable Download Scientific Diagram



Crispr Cas9 Guide Rna Design Rules For Predicting Activity Springerlink



Crispr Cas9 Cleavage Efficiency Correlates Strongly With Target Sgrna Folding Stability From Physical Mechanism To Off Target Assessment Scientific Reports



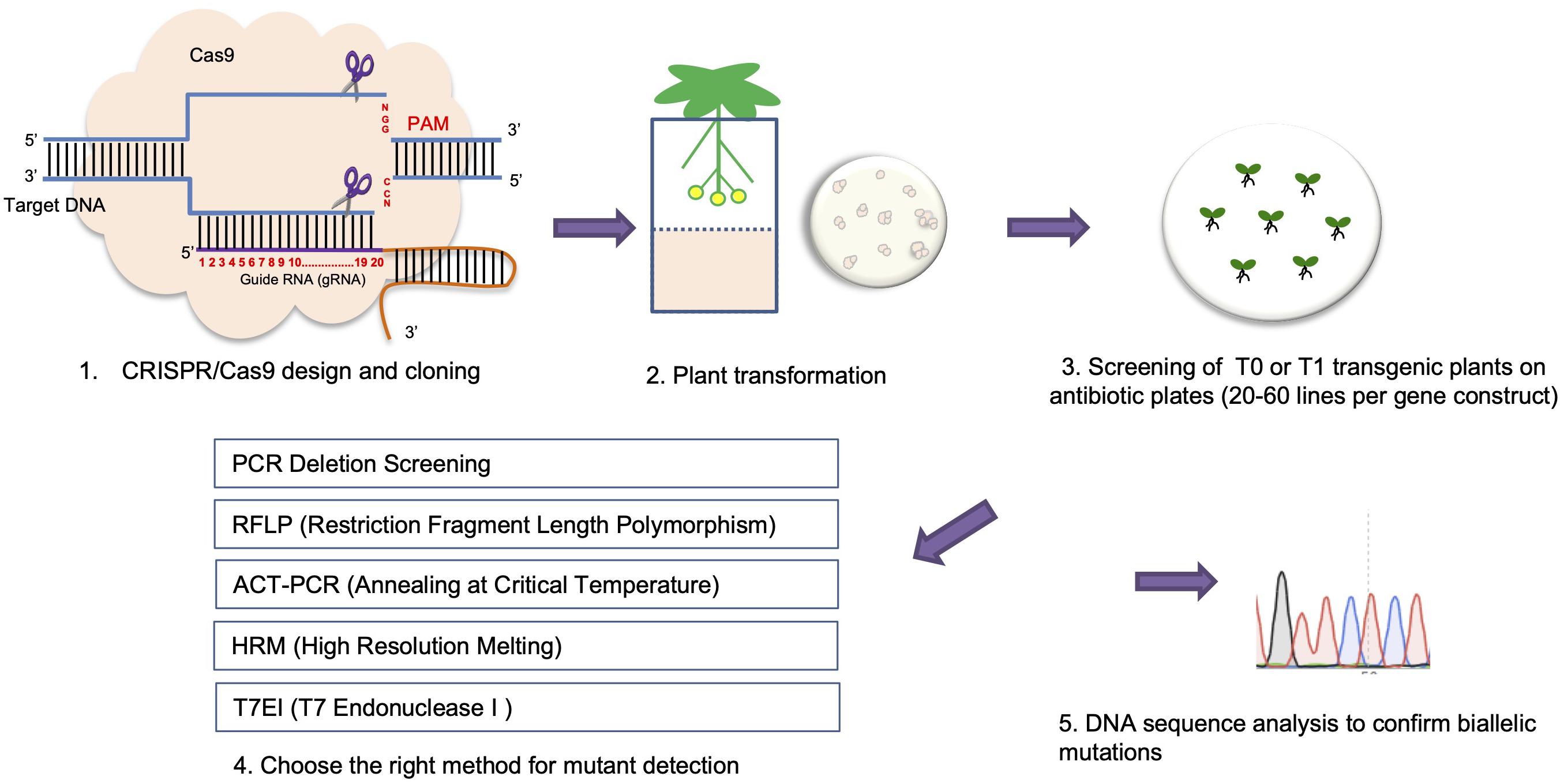
Frontiers Crispr Cas9 Genome Editing Technology A Valuable Tool For Understanding Plant Cell Wall Biosynthesis And Function Plant Science



Crispr Cas9 Guide Rna Specificity




Figure 3 From Crispr Interference Crispri For Sequence Specific Control Of Gene Expression Semantic Scholar




Addgene Crispr Guide
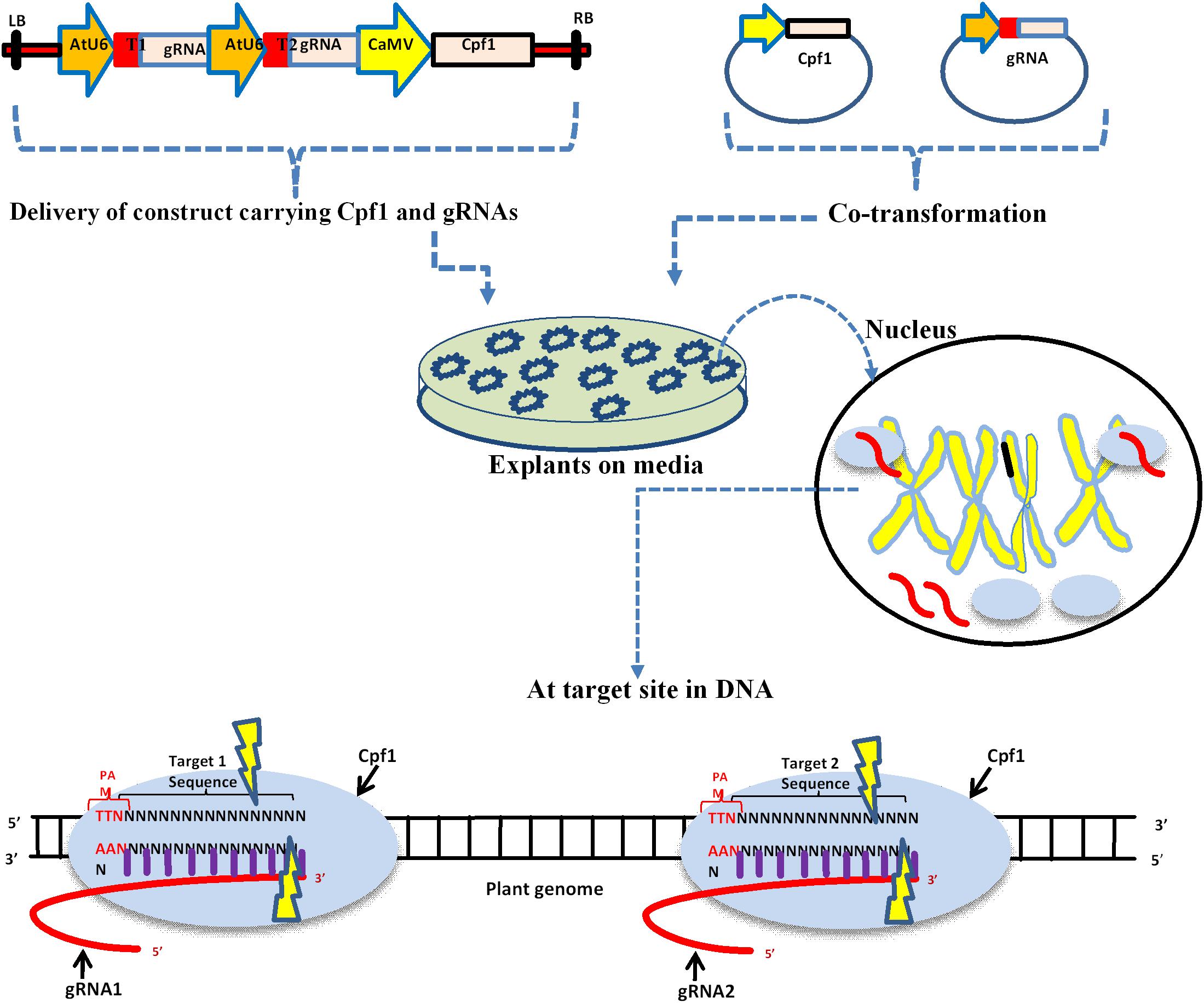



Frontiers The Rise Of The Crispr Cpf1 System For Efficient Genome Editing In Plants Plant Science




Rna Guided Genome Editing In Plants Using A Crispr Cas System Sciencedirect
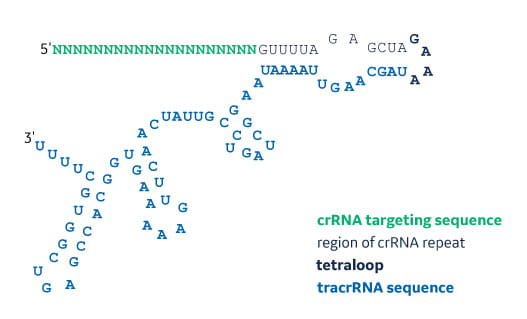



Synthetic Sgrna For Crispr Cas9 Experiments
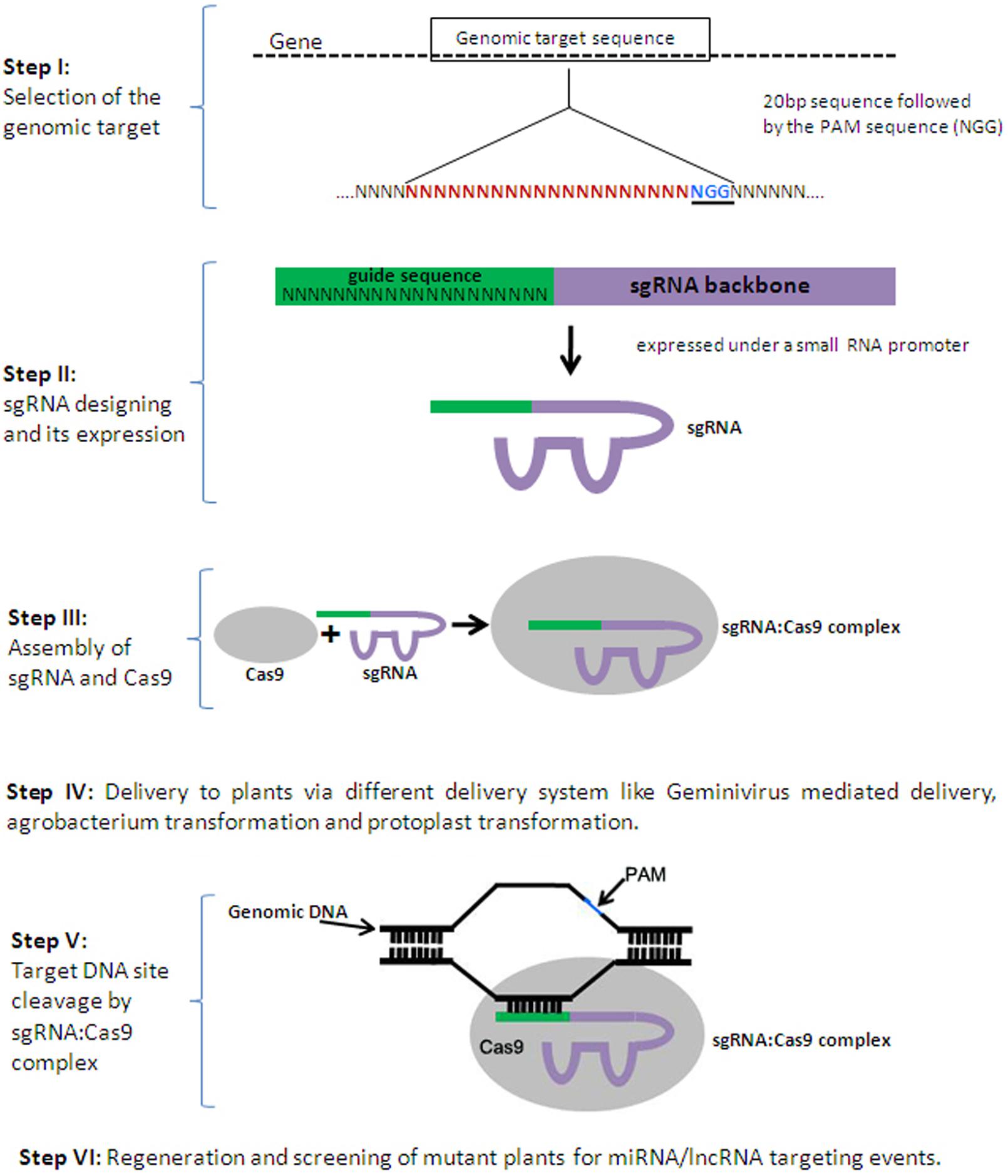



Frontiers Targeting Non Coding Rnas In Plants With The Crispr Cas Technology Is A Challenge Yet Worth Accepting Plant Science
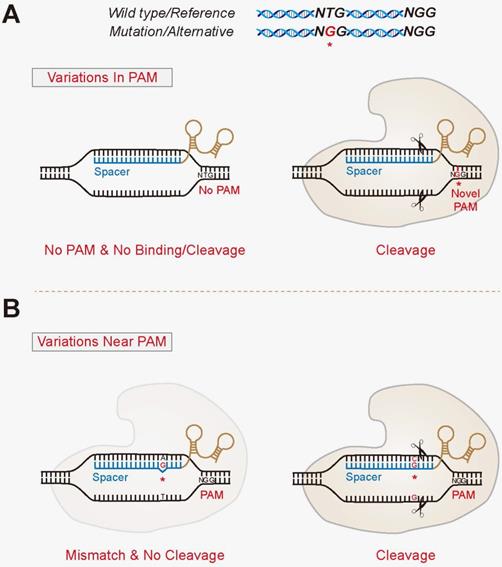



Allele Specific Genome Targeting In The Development Of Precision Medicine




Overview Of The Crispr Cas9 Mechanism Of Action A Crispr Cas Download Scientific Diagram
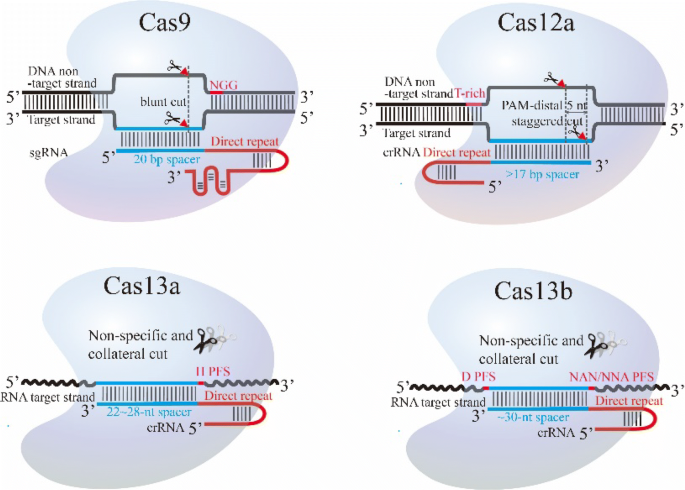



Class 2 Crispr Cas An Expanding Biotechnology Toolbox For And Beyond Genome Editing Cell Bioscience Full Text
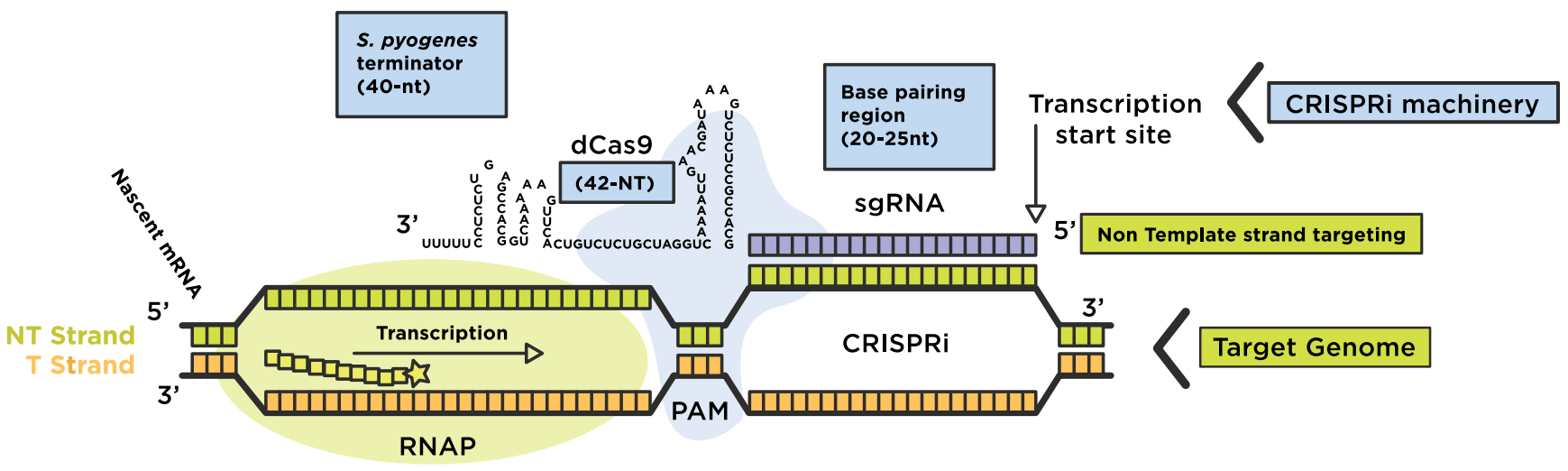



5 Key Steps To Successful Crispri Technology Networks




Truncated Scrrnas Mediate Gene Disruption Comparable To Download Scientific Diagram
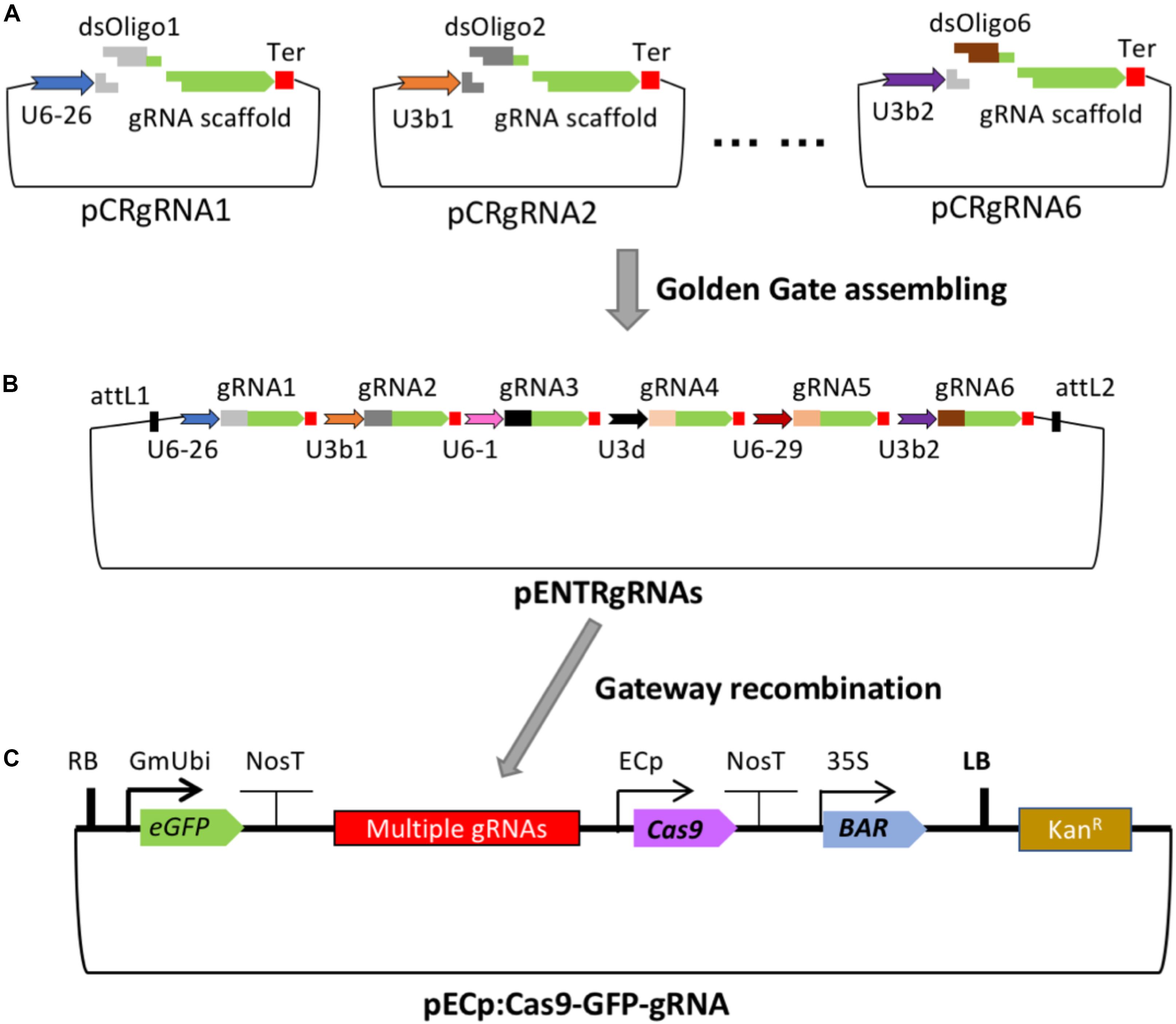



Frontiers Crispr Cas9 Based Gene Editing Using Egg Cell Specific Promoters In Arabidopsis And Soybean Plant Science
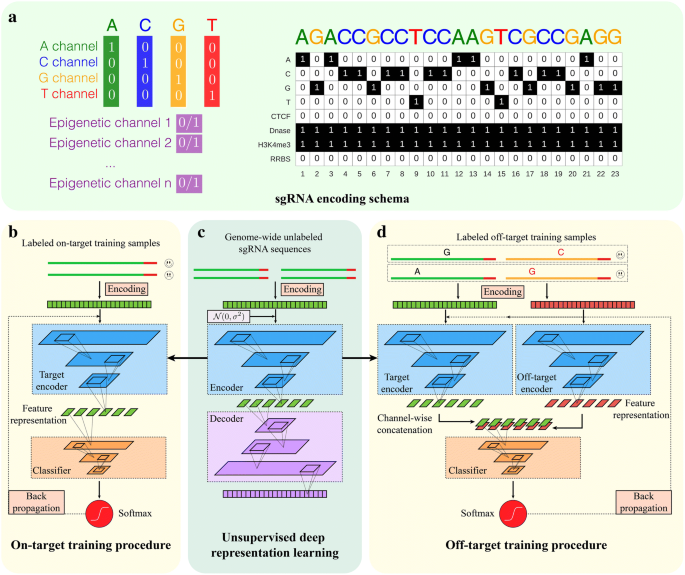



Deepcrispr Optimized Crispr Guide Rna Design By Deep Learning Genome Biology Full Text




The Complete Guide To Understanding Crispr Sgrna




High Throughput Assay For Nicking And Cleavage By Crispr Cas Download Scientific Diagram



Viruses Free Full Text Plant Viruses From Targets To Tools For Crispr Html



Evaluating The Cleavage Efficacy Of Crispr Cas9 Sgrnas Targeting Ineffective Regions Of Arabidopsis Thaliana Genome Peerj
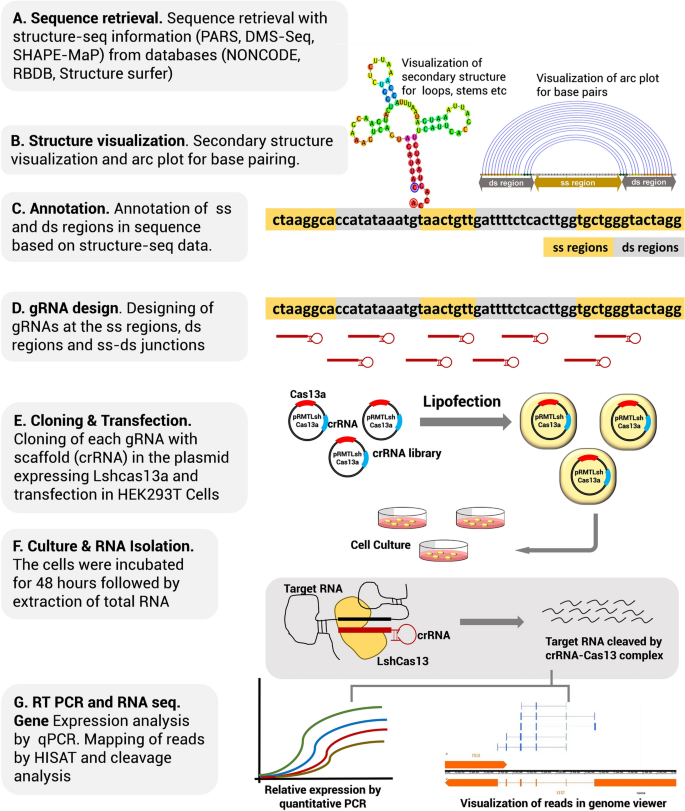



Structure Based Design Of Grna For Cas13 Scientific Reports



2



Team Scu China Project Regulation 18 Igem Org




Addgene Crispr Guide




Harnessing Crispr Cas Systems For Bacterial Genome Editing Trends In Microbiology



Offscan A Universal And Fast Crispr Off Target Sites Detection Tool Bmc Genomics Full Text



A Cas9 Is Directed To A Specific Genomic Target By The First Nt Of Download Scientific Diagram




A 5 Nucleotide Seed For Dcas9 Binding A Most Peaks Are Associated Download Scientific Diagram



1
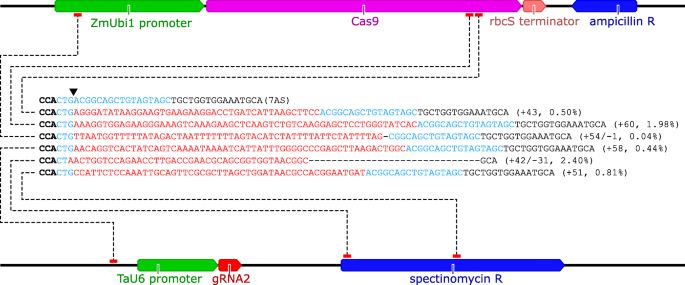



Grna Validation For Wheat Genome Editing With The Crispr Cas9 System Bmc Biotechnology Full Text



2
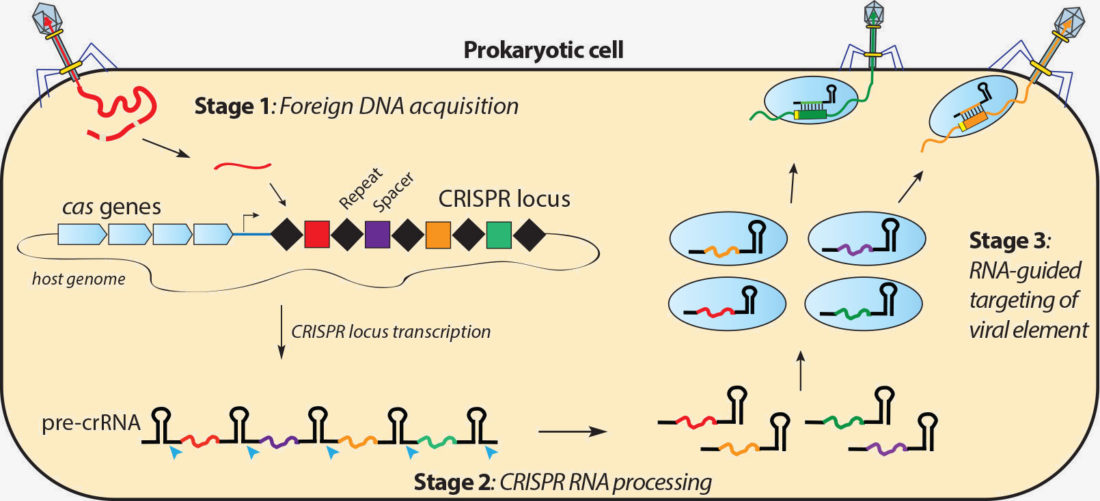



Crispr Systems Doudna Lab
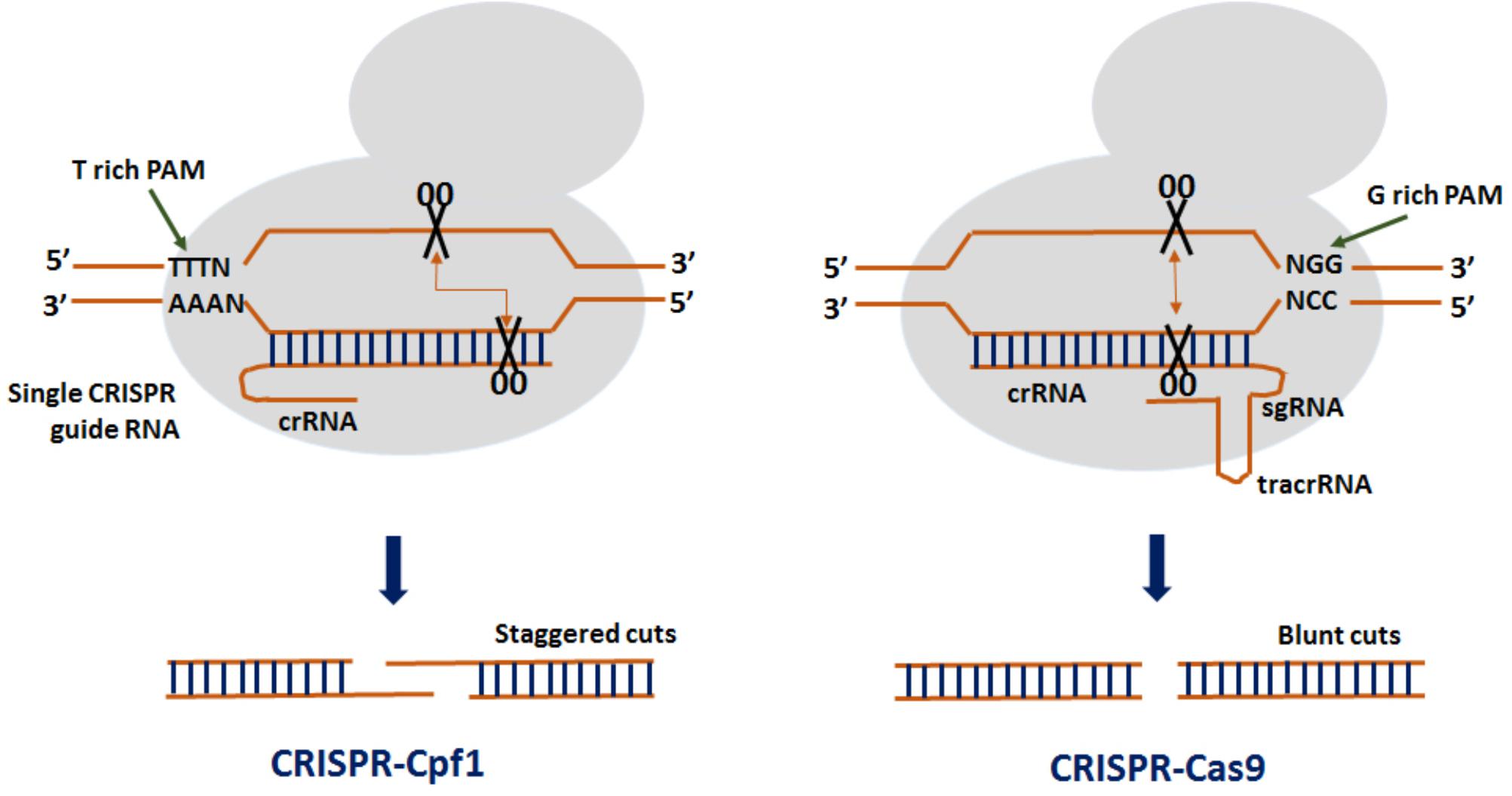



Frontiers Genome Editing In Rice Recent Advances Challenges And Future Implications Plant Science
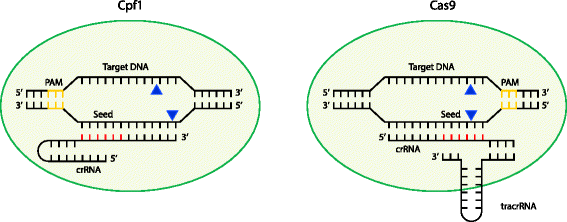



The Cpf1 Crispr Cas Protein Expands Genome Editing Tools Genome Biology Full Text




Base Preferences For Crispr Cas9 System Cas9 Protein Is Shown In Download Scientific Diagram



Plos One Sgrnacas9 A Software Package For Designing Crispr Sgrna And Evaluating Potential Off Target Cleavage Sites




Targeted Transcriptional Repression In Bacteria Using Crispr Interference Crispri Abstract Europe Pmc



Crispr Cpf1 Proteins Structure Function And Implications For Genome Editing Cell Bioscience Full Text



Partial Dna Guided Cas9 Enables Genome Editing With Reduced Off Target Activity Nature Chemical Biology




Off Target Genome Editing Wikipedia



Frontiers Crispr Cas9 Genome Editing Technology A Valuable Tool For Understanding Plant Cell Wall Biosynthesis And Function Plant Science



Frontiers Using Synthetically Engineered Guide Rnas To Enhance Crispr Genome Editing Systems In Mammalian Cells Genome Editing




Systematic Analysis Of Crispr Cas9 Mismatch Tolerance Reveals Low Levels Of Off Target Activity Sciencedirect



1




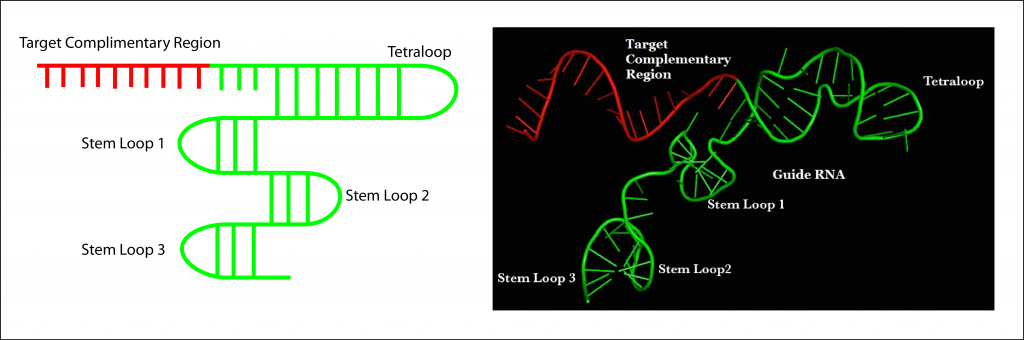
Type Ii Crispr Formulations Grnas Contain 4 Loop Structures Download Scientific Diagram




Design Principle Of A Crispr Cas9 Expression Vector For Construction Download Scientific Diagram



2




Genome Modification By Crispr Cas9 Ma 14 The Febs Journal Wiley Online Library



Crispr Cas9




Sequence Recognition And Structure Of Synthetic Crispr Rna A Download Scientific Diagram



1




The Tolerance Profile Of Sgrna Target Mutation A A Tolerance Profile Download Scientific Diagram




Multiplex And Hdr Mediated Genome Editing By Sgrna Pcocas9 In Plant Download Scientific Diagram




Crispr Cas Systems In Genome Editing Methodologies And Tools For Sgrna Design Off Target Evaluation And Strategies To Mitigate Off Target Effects Manghwar Advanced Science Wiley Online Library




Dual Sgrna Based Targeted Deletion Of Large Genomic Regions




One Or Two Mismatches In The Seed Can Be Tolerated For Crispr Activity Download Scientific Diagram
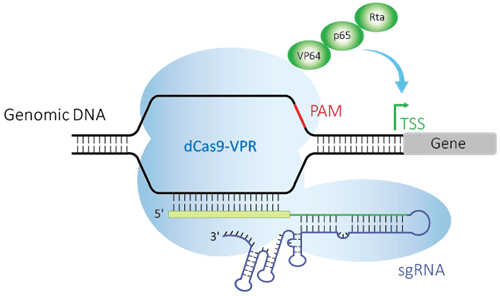



Crispr Activation




Nucleosomes Selectively Inhibit Cas9 Off Target Activity At A Site Located At The Nucleosome Edge Journal Of Biological Chemistry




Sgrna Sequence Motifs Blocking Efficient Crispr Cas9 Mediated Gene Editing Sciencedirect



Crispr




Addgene Crispr Guide
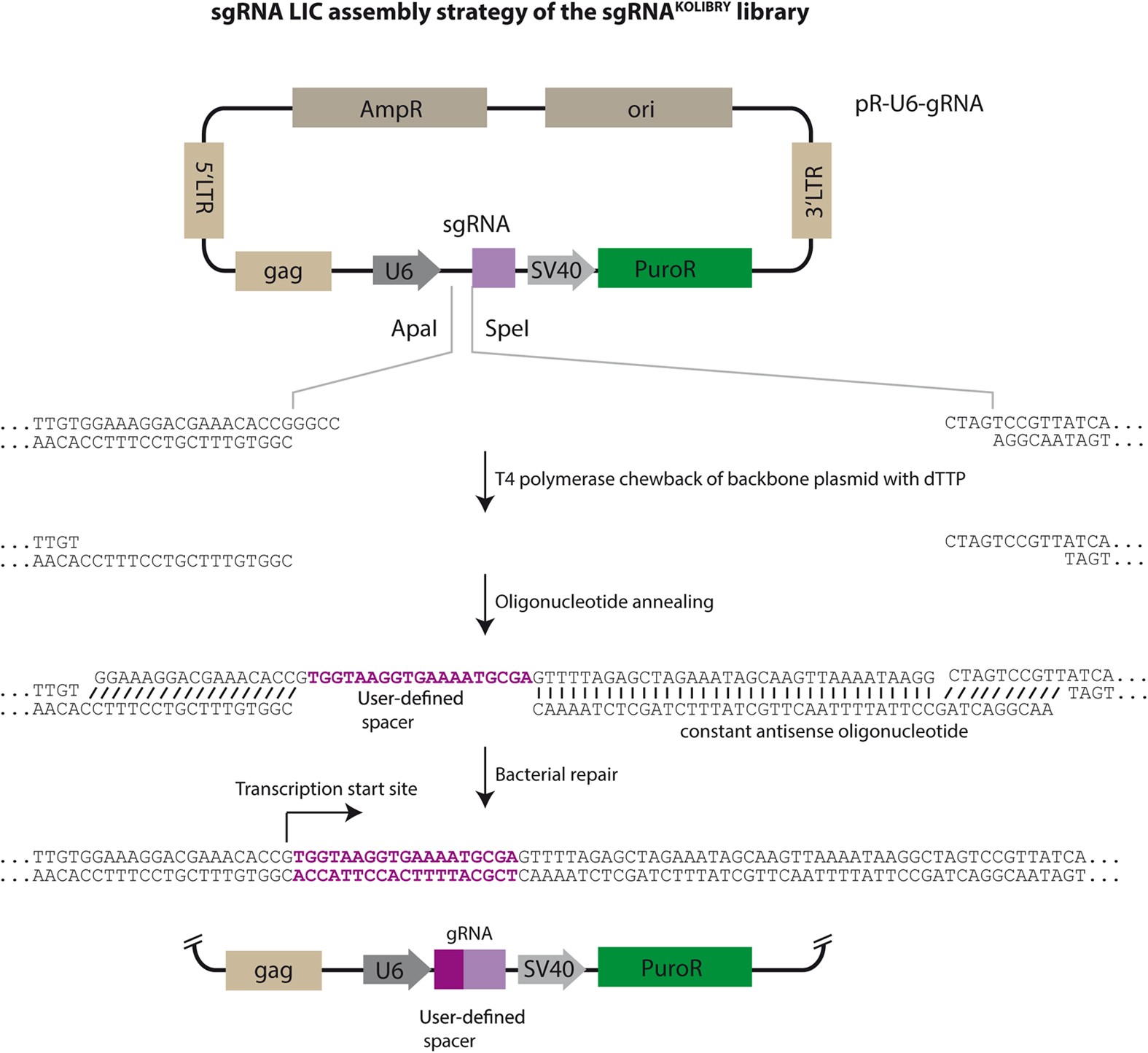



Synthesis Of An Arrayed Sgrna Library Targeting The Human Genome Scientific Reports



Cas9 Mechanism Crispr Cas9



Tsl Synbio
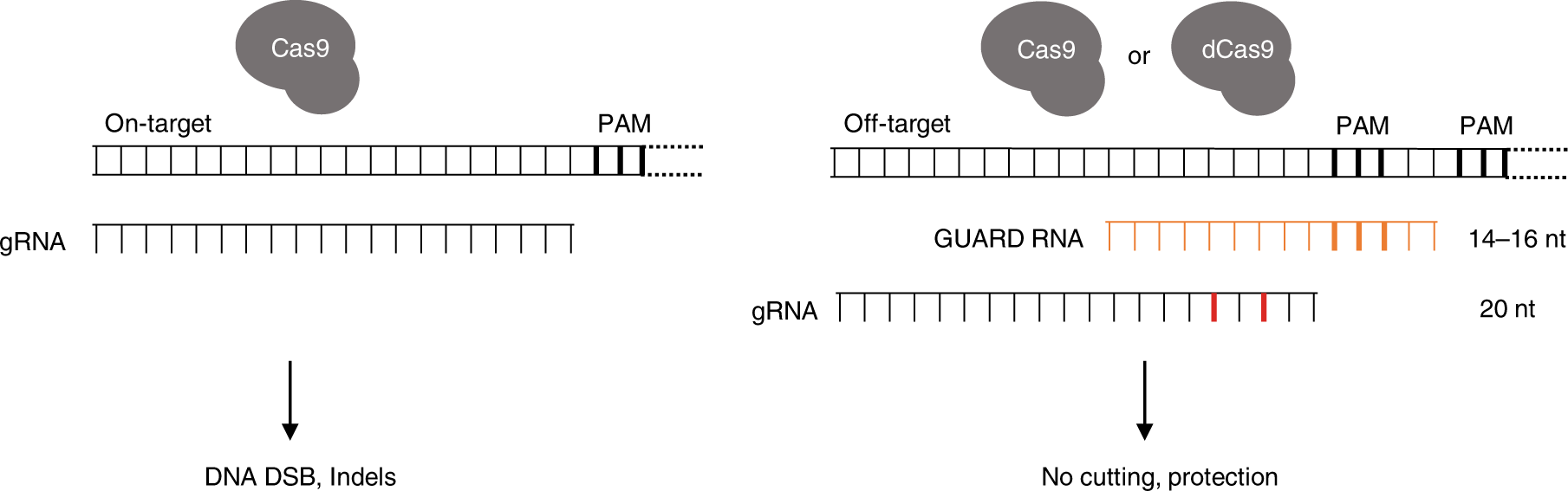



Crispr Guard Protects Off Target Sites From Cas9 Nuclease Activity Using Short Guide Rnas Nature Communications
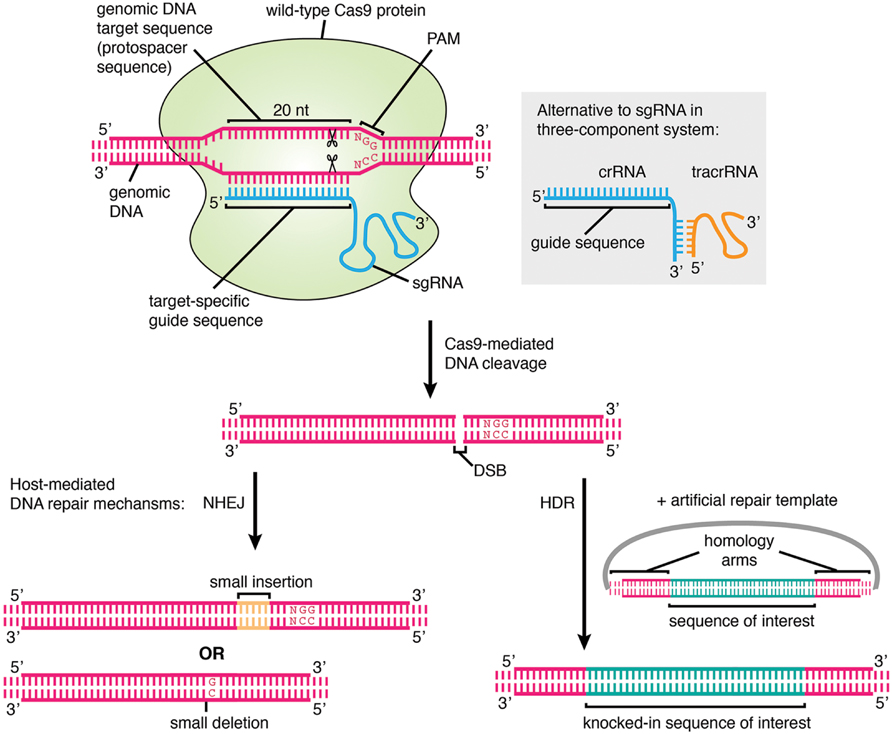



Frontiers A New Age In Functional Genomics Using Crispr Cas9 In Arrayed Library Screening Genetics




Overview Of Guide Rna Design Tools For Crispr Cas9 Genome Editing Technology
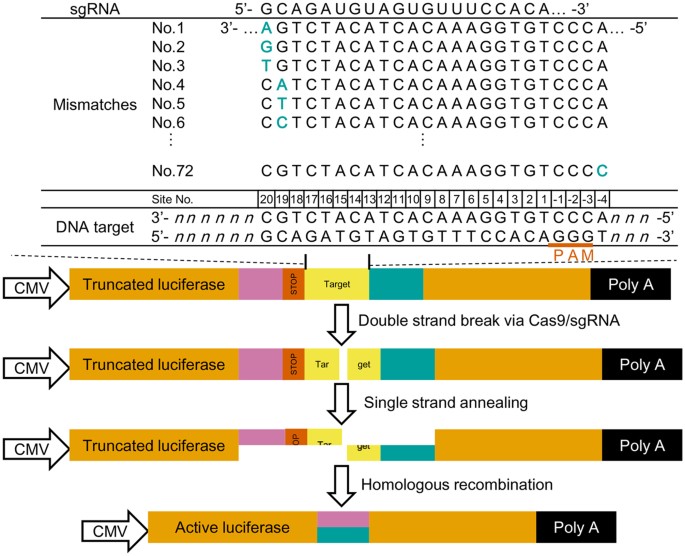



Profiling Single Guide Rna Specificity Reveals A Mismatch Sensitive Core Sequence Scientific Reports




Crispr Cas9 Directed Gene Editing For The Generation Of Loss Of Function Mutants In High Throughput Zebrafish F0 Screens Shankaran 17 Current Protocols In Molecular Biology Wiley Online Library




Computational Approaches For Effective Crispr Guide Rna Design And Evaluation Sciencedirect




A Crispr Cas9 Triggered Strand Displacement Amplification Method For Ultrasensitive Dna Detection Nature Communications
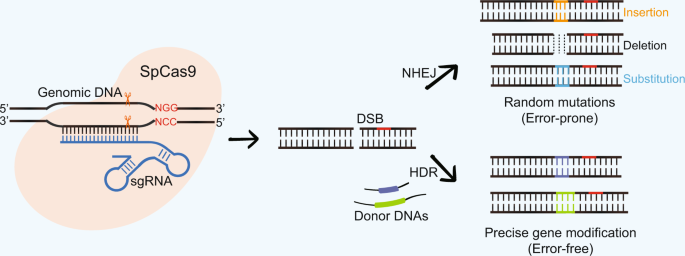



Crispr Technology Is Revolutionizing The Improvement Of Tomato And Other Fruit Crops Horticulture Research




Efficient Multiplex Biallelic Zebrafish Genome Editing Using A Crispr Nuclease System Pnas




Versatile Modification Of The Crispr Cas9 Ribonucleoprotein System To Facilitate In Vivo Application Sciencedirect



2




Evaluation Of Off Targets Predicted By Sgrna Design Tools Sciencedirect




Sample Input File And Possible Off Target Types In The Casot Download Scientific Diagram
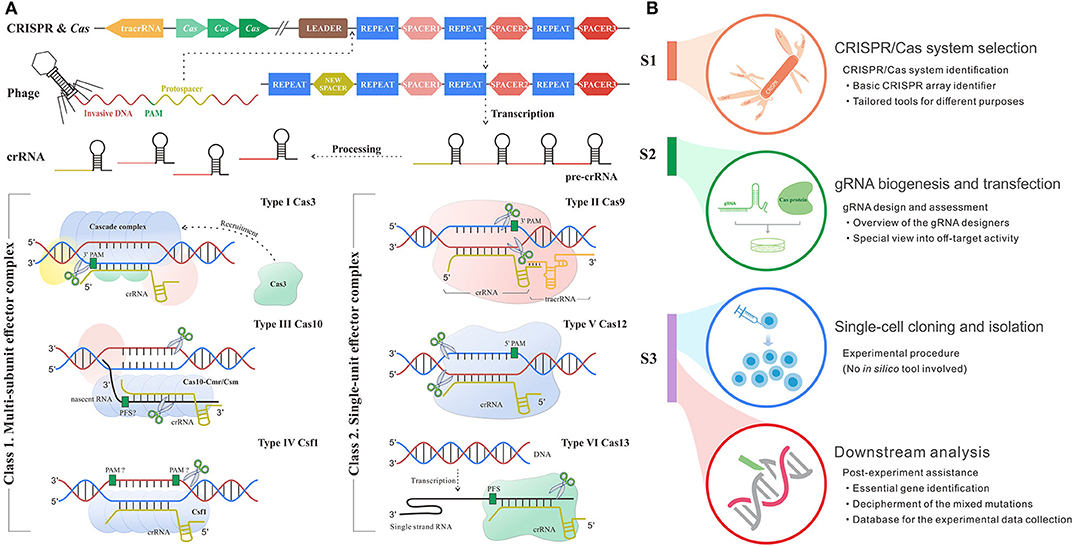



Frontiers In Silico Method In Crispr Cas System An Expedite And Powerful Booster Oncology




Binding Of Central Seed Region In Grna Crucial For Cas13 Mediated Download Scientific Diagram




Off Target Effects In Crispr Cas9 Mediated Genome Engineering Sciencedirect



0 件のコメント:
コメントを投稿